Using the function FILES to select files for fMRI data analysis
File selection is one of the most time consuming aspects of working with the SPM GUI. More importantly, having to manually select files in order to process your data makes it virtually impossible to automate your analysis pipeline. But you're in luck: You are in possession of a MATLAB function called "FILES" that, with just one line of code from the command line, will grab all the files that you're looking for. Here, I illustrate how files can be used to grab a specific file from all 10 subjects. In addition, I illustrate why this is useful with a function called BSPM_CHECKREG (which is basically a command line version of the "Check Reg" button you're already familiar with).
Contents
Use the HELP function to see documentation for using FILES
help files
FILES OS-Sensitive Wrapper for RDIR
USAGE: [fullpath, filename] = files(pattern, [celloutput])
(adapted from RDIR documentation) "pattern" can be a pathname,
filename, or can include both. One can use absolute and relative
pathnames and wildcards (*). Wildcard can be placed anywhere and used
many times like 'path*\*.m'. One can also use a double wildcard (**) to
match multiple directory levels. For example pattern = 'path\**\*.m'
will match all ".m" files in "path" and all subdirectories of "path".
INPUT
pattern: string to use to select files (wildcards OK)
charoutput: output as cell arrays [0=no (default), 1 = yes]
OUTPUTS
fullpath: full path to filenames (char by default)
filename: filename only (cekl array by default) (OPTIONAL)
This is a wrapper function for RDIR.m, which is included in this file
as a subfunction. RDIR.m was downloaded from the MATLAB File Exchange:
www.mathworks.com/matlabcentral/fileexchange/
32226-recursive-directory-listing-enhanced-rdir
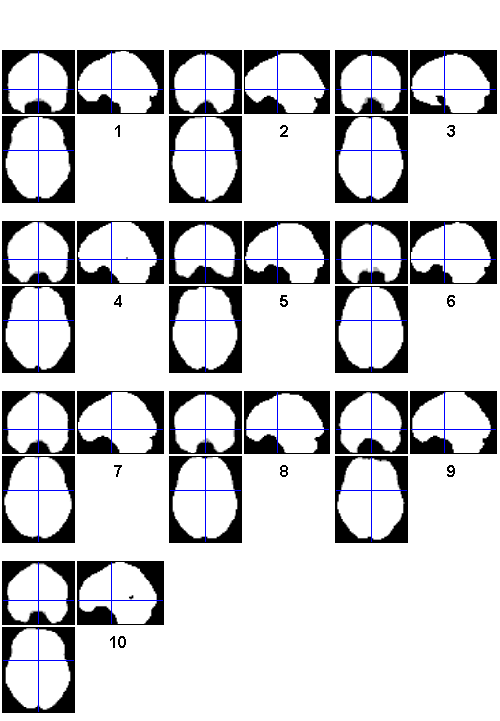
Use FILES to select all 10 mask.img files
allmask = files('sub*/**/precooked/2x2/mask.img', 1)
allmask = /Users/bobspunt/Desktop/Personal/SPM_Labs_Data/sub01/results/precooked/2x2/mask.img /Users/bobspunt/Desktop/Personal/SPM_Labs_Data/sub02/results/precooked/2x2/mask.img /Users/bobspunt/Desktop/Personal/SPM_Labs_Data/sub03/results/precooked/2x2/mask.img /Users/bobspunt/Desktop/Personal/SPM_Labs_Data/sub04/results/precooked/2x2/mask.img /Users/bobspunt/Desktop/Personal/SPM_Labs_Data/sub05/results/precooked/2x2/mask.img /Users/bobspunt/Desktop/Personal/SPM_Labs_Data/sub06/results/precooked/2x2/mask.img /Users/bobspunt/Desktop/Personal/SPM_Labs_Data/sub07/results/precooked/2x2/mask.img /Users/bobspunt/Desktop/Personal/SPM_Labs_Data/sub08/results/precooked/2x2/mask.img /Users/bobspunt/Desktop/Personal/SPM_Labs_Data/sub09/results/precooked/2x2/mask.img /Users/bobspunt/Desktop/Personal/SPM_Labs_Data/sub10/results/precooked/2x2/mask.img
Use BSPM_CHECKREG to examine the selected images using Check Reg
bspm_checkreg(allmask);
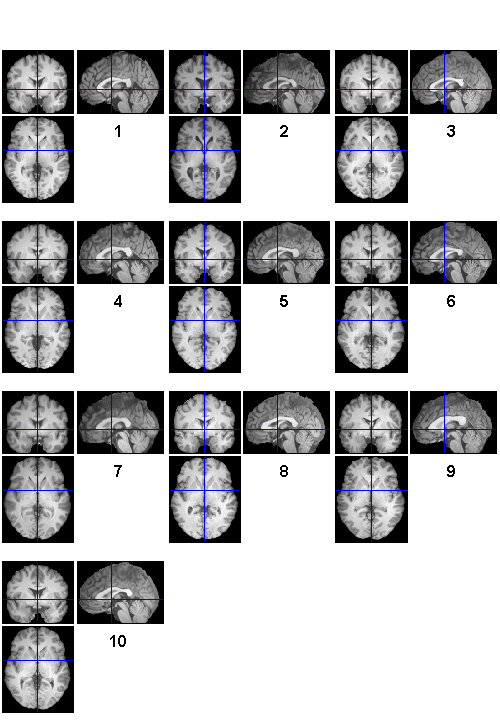
Use FILES to select all 10 wanat_hires.nii files
allanat = files('sub*/**/wanat_hires.nii', 1)
allanat = /Users/bobspunt/Desktop/Personal/SPM_Labs_Data/sub01/anatomy/precooked/wanat_hires.nii /Users/bobspunt/Desktop/Personal/SPM_Labs_Data/sub02/anatomy/precooked/wanat_hires.nii /Users/bobspunt/Desktop/Personal/SPM_Labs_Data/sub03/anatomy/precooked/wanat_hires.nii /Users/bobspunt/Desktop/Personal/SPM_Labs_Data/sub04/anatomy/precooked/wanat_hires.nii /Users/bobspunt/Desktop/Personal/SPM_Labs_Data/sub05/anatomy/precooked/wanat_hires.nii /Users/bobspunt/Desktop/Personal/SPM_Labs_Data/sub06/anatomy/precooked/wanat_hires.nii /Users/bobspunt/Desktop/Personal/SPM_Labs_Data/sub07/anatomy/precooked/wanat_hires.nii /Users/bobspunt/Desktop/Personal/SPM_Labs_Data/sub08/anatomy/precooked/wanat_hires.nii /Users/bobspunt/Desktop/Personal/SPM_Labs_Data/sub09/anatomy/precooked/wanat_hires.nii /Users/bobspunt/Desktop/Personal/SPM_Labs_Data/sub10/anatomy/precooked/wanat_hires.nii
Use BSPM_CHECKREG to examine the selected images using Check Reg
bspm_checkreg(allanat);